Table of Contents
Challenges in proteomics
In the past several decades researchers have had to rely on mass spectrometry-based techniques to quantify and identify the proteins involved in their samples. These techniques were low throughput however and often required difficult protocol optimizations and time-consuming data analysis pipelines. While useful, these techniques were inadequately slow and required a lot of “hands on” time from researchers. It wouldn’t be long before a new approach to studying proteomics was discovered, using protein chips or microarrays researchers were able to facilitate large-scale, high-throughput proteomic studies. Despite aiding in the advancement of the field, analyzing the data and delineating protein concentrations and interactions from large-scale datasets remained challenging.
A new method of studying protein interactions
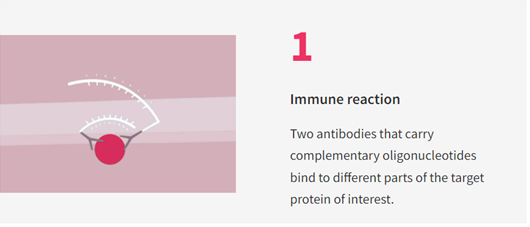
Olink’s Proximity Extension Assay (PEA) technology is a new method of quantifying and identifying proteins of interest in complex samples such as blood serum or plasma. Olink platforms such as the Explore HT® and Explore 3072® uses PCR amplification of DNA tags unique to each target protein in proportion to the abundance of the target proteins in the sample: First, two antibodies that carry complementary oligonucleotides (acting as unique barcodes for each antibody) bind to different parts of the target protein must be brought close to each other on the target protein (see Figure 1).
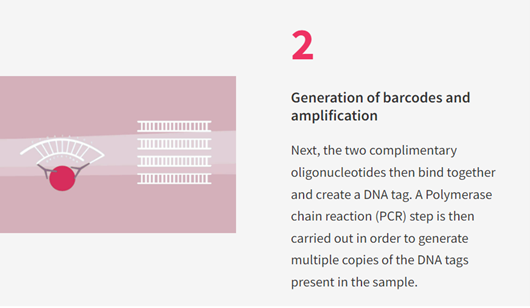
When successful PCR amplification of a DNA tag for the antibody target protein occurs, quantitative PCR reports it as a proxy for the abundance of the target protein (see Figure 2).
Finally, a short-read Next Generation Sequencing (NGS) platform is used to measure the abundance of the target protein by measuring the proportion of the DNA tags present after PCR (see Figure 3).

With this novel approach to biomarker discovery, researchers are now able to utilize Olink’s HT and 3072 kits in combination with liquid handling and short read sequencing platforms to identify/quantify low abundance proteins with high specificity and accuracy in serum or plasma samples.
A new approach to biomarker discovery
The first step in the workflow involves selecting either the Olink HT or 3072 kit to measure over 5300+ proteins or 3000 proteins, respectively, across 10 orders of magnitude in protein concentrations. Here, Olink’s industry-leading specificity is founded on PEA technology (me, which generates signals only when two matched oligonucleotide-tagged antibodies bind to their intended target protein and generate a correctly hybridized DNA template for readout, eliminating false positives and ensuring data accuracy.
Explore HT workflow
- Sample Preparation & Dilution using liquid handling platforms such as those manufactured by Hamilton and the Mosquito/Dragonfly platforms by SPT Labtech
- Sample incubation prior to PCR step with the Mosquito liquid handling platform
- PCR sets up with Mosquito and Dragonfly, and PCR pooling steps using Sciclone G3 NGSx liquid handling platforms.
- A manual library purification step along with Quality Control
- The NGS libraries are then sequenced on Illumina’s NovaSeq 6000 short read sequencing platform
- Data analysis of results using Olink’s proprietary analysis software. Automation reduces manual steps, while excluding data points that don’t meet criteria before normalization. Moreover, the software effectively identifies the underlying causes of any issues, pinpointing technical and human errors.
- Olink Pre-Process software ngs2counts
- Olink NPX Explore HT Software
Explore 3072 workflow
- Sample Preparation & Dilution using liquid handling platforms such as those manufactured by Hamilton and the Mosquito/Dragonfly platforms by SPT Labtech
- Incubation preparation with the Mosquito platform
- PCR 1 sets up with Dragonfly, PCR 1 pooling and PCR 2 sets up, pooling steps with Sciclone G3 NGSx platforms
- Manual Library Purification steps along with Quality Control
- The NGS libraries are then sequenced on Illumina’s NovaSeq 6000 short read sequencing platform
- Data analysis of results using Olink’s proprietary analysis software. Automation reduces manual steps, while excluding data points that don’t meet criteria before normalization. Moreover, the software effectively identifies the underlying causes of any issues, pinpointing technical and human errors.
- Olink Pre-Process software ngs2counts
- Olink NPX Explore Software
Overcoming the challenges in high-throughput biomarker discovery
Olink’s Explore HT and 3072 kits are just one part of the biomarker discovery process, as shown in the workflow above, liquid handling platforms such as SPT Labtech’s Dragonfly instruments are also necessary in order to accurately add dilutant and PCR master mix. Briefly, the Dragonfly platform harnesses innovative positive displacement technology to enable accurate and repeatable nanoliter to milliliter dispensing, every time to ensure accuracy and repeatability. While the Mosquito dilutes samples and adds assay reagents for both Olink kits. The Mosquito also utilizes positive displacement pipetting solutions and combines this with the power of miniaturization and automation to reduce costs, improve productivity, and maximize research output.
Once the PCR step has concluded and the NGS libraries are prepared, the library can be sequenced using Illumina’s NovaSeq 6000 platform. The NovaSeq 6000 is a powerful sequencer that can generate massive amounts of short-read sequencing data in a short period of time. The sequencing data is processed using bioinformatics tools to align the reads to a reference genome, identify single-nucleotide polymorphisms (SNPs) and quantify gene expression levels.
Finally, trained experts are then able to take the raw data taken from the NovaSeq 6000 platform and normalize it using Olink’s proprietary software to yield “Normalized protein expression” values or NPX.
In conclusion, the Olink Explore HT/3072 kits, SPT Labtech liquid handling along with Hamilton and Sciclone G3 NGSx platforms and Illumina’s NovaSeq 6000 platform provide a comprehensive workflow for high-throughput biomarker discovery. This workflow enables researchers to quickly and accurately identify potential biomarkers that can be used for diagnostic or therapeutic purposes. However, purchasing, maintaining, and training staff to run these platforms is highly expensive and time consuming.
Outsourcing biomarker discovery to Sampled
There is an alternative to purchasing these platforms and kits. Outsourcing your samples for high throughput biomarker discovery to Sampled allows researchers to save on capital expenditure since you no longer need to invest in platforms, consumables, and reagents for each step in this workflow outlines above. Furthermore, outsourcing removes the need to train staff on new platforms and time-consuming optimization steps required for each step of the workflow. Our trained experts have been trained by each of these manufacturers allowing us to provide industry-leading turnaround time and accuracy.
Speak to one of our experts today to learn more about how our Protein Biomarker Discovery workflow can accelerate your research.

